Current Projects
BioPoplar

The goal of this project is to re-engineer poplar, one of the fastest growing trees in the United States, as a multipurpose crop that can be used for bioenergy, biomaterial, and bioproduct production as the need for sustainable sources of bioproducts and plant-based materials continues to rise.
more infoConstruction of a Minimal Plant Genome

In this project, we will develop a gene editing platform at scale that will be used to generate novel plant genotypes with reduced gene content and redundancy, which can serve as chassis (i.e., a platform) to rapidly engineer plants with new phenotypes and test heterologous genes relevant to biological processes of interest.
more infoEngineering a terpene-depleted chassis in tomato fruit for production of high value terpenes

The goal of this project is to eliminate the factors required for making native triterpenes and carotenoids in fruit to allow expression of biosynthetic pathways from other organisms for sustainable production of high value terpenes.
more infoGenome-enabled breeding across Phaseolus species

Phaseolus vulgaris, common or dry bean, diverged from its sister species, tepary bean (Phaseolus acutifolius) 2.1 MYA. We will transform common and tepary bean breeding by exploiting their close evolutionary relationship with the development of a computational genomics framework that permits rapid interspecific trait discovery and subsequent introgression using bridging genotypes.
more infoSingle cell-enabled genome-to-pathway discovery of plant natural product biosynthetic pathways

Plants produce a plethora of medicinally important natural products that have significant potential to improve human health yet are often produced in low amounts, in complex mixtures, and/or cannot be produced in a sustainable or economic manner. Omics technologies enable the discovery of the biosynthetic genes of plant natural products which can then be employed in commercial production of medically relevant compounds. In this project, we will use state-of-the-art omics technologies to efficiently discover genes responsible for the biosynthesis of pharmacologically relevant compounds and utilize this knowledge to revitalize an old technology, plant cell culture, to enable robust, sustainable production of these products.
more infoWheat Diversity Project

Wheat is a major crop providing ~20% of dietary calories and proteins worldwide, and a major concern for global food security. Development of the initial International Wheat Genome Sequencing Consortium Chinese Spring RefSeq served as a springboard for genetics, evolution, breeding and biotechnology advancements in wheat. In this project, we will update the Chinese Spring genome assembly and provide new landrace genomes to enable further advancements in genome-enable research in wheat.
more infoFormer Projects
Accelerated Resistance Gene Discovery in Common Bean
A USDA funded project to enhance durable disease resistance in beans using genomics-enabled plant breeding techniques.
more infoBiofuel Feedstock Genomics Resource

The Biofuel Feedstock Genomics Resource is a database that provides fully integrated and uniform annotation to a large collection of plant species that are being examined for possible use as ligno-cellulosic feedstocks for biofuel production.
Blueberry Medicinal Genomics

A NIH-funded project to investigate the biosynthesis of iridoid compounds in blueberry, a class of pharmacologically important compounds that have positive human health benefits.
more infoCircadian Clock in Potato
This project is focused on the role of the circadian clock in potato domestication and development through the use of genetic, genomic, cell/molecular biology, and bioinformatic techniques.
more infoClimate Change and Potato

As part of the MSU Project GREEEN we are investigating how potatoes produced in Michigan will be impacted by altered CO2 and temperature conditions anticipated in the mid-21st century.
more infoCold Tolerance in Switchgrass

As part of the DOE USDA Feedstock Genomics for Bioenergy Project and in collaboration with USDA-ARS, we are examining genomic variations from exome capture and transcriptome sequencing of switchgrass populations to improve the efficiency of breeding switchgrass cultivars with high biomass and cold hardiness.
more infoComprehensive Phytopathogen Genomics Resource

The Comprehensive Phytopathogen Genomics Resource (CPGR) is a comprehensive plant pathogen genomics and annotation resource funded by USDA CSREES. A focus of the project is the development of plant pathogen diagnostic markers using genomics data.
more infoEffects of Root and Shoot Architecture on Nutrition in Dry Bean

This project aims to identify indirect selection events in dry bean root systems after 100+ years of breeding for shoot architecture traits.
more infoEvolutionary Origins of Tubers
Major crop species use tubers to bypass sexual reproduction. This project aims to identify how tubers have evolved multiple times and provide insight into tuber development.
more infoGenome wide evaluation of off-targets from gene editing reagents in seed vs. vegetatively propagated crop species

The overall objective of this project is to address genome-wide variability/risk assessment of current genome-editing technology involving CRISPR systems, TALENs and base editors.
more infoGenome Editing

The recent evolution of genome editing technologies has transformed the field of plant biotechnology and provided the opportunity for crop improvement. With funding from USDA-Biotechnology Risk Assessment Grants Program (BRAG) we explore the impact of genome editing technologies in potato at the whole genome and transcriptome levels.
more infoGenomic Tools for Sweetpotato (GT4SP) Improvement Project

We are participating in a consortium led by North Carolina State University funded by the Bill & Melinda Gates Foundation whose primary aims are to develop new genomic, genetic, and bioinformatics tools for improvement of hexaploid sweetpotato. See Sweetpotato Genomics Resource and the GT4SP website for more information.
more infoMaize Genetic Diversity

As part of the Great Lakes Bioenergy Research Center, we are examining natural diversity in maize and associating genes and alleles with traits important to use of stover in biofuel feedstock production.
more infoMaize Genomics Resource (MGR) Annotation Project

The Maize Genome Resource provides a range of genomic tools for maize B73. A notable feature is the EFP browser for the developmental B73 gene atlas and several B73 abiotic and biotic stress experiments that were published in Hoopes et al (2018). The MGR also features a comprehensive and regularly updated JBrowse instance including a range of annotation tracks from published data sets.
more infoMaize Nutrigenomics

We are participating in a collaborative project funded through the National Science Foundation Plant Genome Research Program to determine the genetic architecture of vitamin biosynthesis in maize.
Medicinal Plant Genomics

Building upon past accomplishments from our Medicinal Plant Transcriptomics project we have continued our work on generating genomic resources and functional annotation of several medicinal plants with the target of functionally characterizing genes involved in natural product biosynthesis and facilitating the artificial synthesis of economically pivotal specialized metabolites in heterologous organisms.
Medicinal Plant Transcriptomics

This project, funded by the NIH, addresses the gap in our knowledge base of species-specific plant metabolism. Data generated through this project will accelerate the identification and functional analysis of genes involved in natural product biosynthesis in 14 widely used medicinal plant species.
Mint Genomics

A NSF-funded project on evolution of chemical diversity in the mint family started in collaboration with scientists at the University of Florida, Purdue University, and the John Innes Centre.
Oryza SNP Project

As part of an international collaboration, we have been involved in the discovery and annotation of SNPs in the rice genome. This project is funded by USDA CSREES.
Pack-MULEs and Genome Evolution in Rice
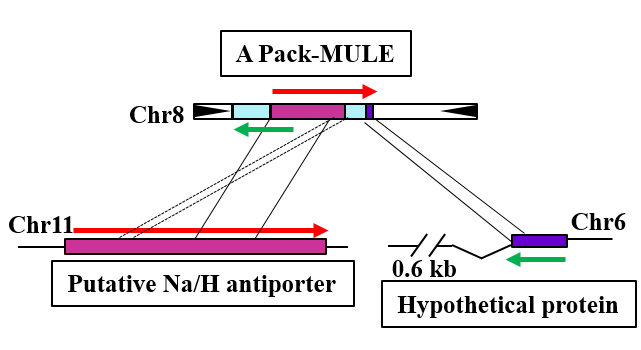
Pack-MULEs are transposable elements that carry fragments of normal protein-coding genes. An NSF funding supports a project on the impact of Pack-MULEs on plant genome evolution and mechanisms of sequence acquisition by Pack-MULEs.
more infoPlant Repeat Database

We have created the Plant Repeat Database to assist in the compilation and identification of repeat sequences in plant genomes.
Plant Resilience Institute

As part of the MSU Plant Resilience Institute, we will be interrogating the genome of tepary bean (Phaseolus acutifolius), a species native to the southwestern U.S. and extremely heat tolerant.
more infoPotato 2.0

Potato is an asexually propagated, cross-pollinated tetraploid crop for which breeding methods have not changed substantially in over 100 years. Current methods for generating new potato cultivars are genetically inefficient due to polyploidy, resource intensive due to vegetative propagation, and do not take full advantage of genomics resources. This project is focused on developing potato as a diploid, inbred-hybrid crop propagated by true seed.
more infoPotato Genome Variation

We are continuing work on an NSF-funded project to understand the contribution of intra-genome variation to potato fitness through the study of tetraploid, diploid, and dihaploid potato.
more infoPythium Genomics
We have been funded by the U.S. Department of Agriculture to sequence and annotate two strains of Pythium ultimum, an important plant pathogen.
Rice Genome Annotation Project

The Rice Genome Annotation Project is an NSF funded effort to provide high quality annotation of the rice genome to the research community.
more infoSingle Cell RNA Sequencing in Plants
Solanaceae Coordinated Agricultural Project

In this collaborative project, we are using high throughput sequencing and bioinformatics to identify variation in elite potato and tomato germplasm for the identification of candidate genes of high value traits such as carbohydrate, sugar and vitamin content. These resources will ultimately be used to develop improved varieties through genotyping and phenotyping panels to associate genes with phenotypes.
SweetGAINS (Genetic Advances and Innovative Seed Systems)

Key to genome-enabled breeding is access to a high quality, annotated genome assembly. We will be annotating the hexaploid Beauregard genome using Oxford Nanopore Technology full length cDNA, mRNAseq data, protein evidence, and gene finders.
more infoSwitchgrass Genetic Diversity

As part of the Great Lakes Bioenergy Research Center, we are examining natural diversity in swithgrass to enable improved breeding efforts for biofuel feedstock production.
more info- BIOPOPLAR
- CONSTRUCTION OF A MINIMAL PLANT GENOME
- ENGINEERING A TERPENE-DEPLETED CHASSIS IN TOMATO FRUIT FOR PRODUCTION OF HIGH VALUE TERPENES
- GENOME-ENABLED BREEDING ACROSS PHASEOLUS SPECIES
- SINGLE CELL-ENABLED GENOME-TO-PATHWAY DISCOVERY OF PLANT NATURAL PRODUCT BIOSYNTHETIC PATHWAYS
- WHEAT DIVERSITY
- ACCELERATED RESISTANCE GENE DISCOVERY IN COMMON BEAN
- BIOFUEL FEEDSTOCK GENOMICS RESOURCE
- BLUEBERRY MEDICINAL GENOMICS
- CIRCADIAN CLOCK IN POTATO
- CLIMATE CHANGE AND POTATO
- COLD TOLERANCE IN SWITCHGRASS
- COMPREHENSIVE PHYTOPATHOGEN GENOMICS RESOURCE
- EFFECTS OF ROOT/SHOOT ARCHITECTURE ON NUTRITION IN DRY BEAN
- EVOLUTIONARY ORIGINS OF TUBERS
- GENE EDITING PROJECT
- GENOME EDITING
- GT4SP IMPROVEMENT PROJECT
- MAIZE GENETIC DIVERSITY
- MAIZE GENOMICS RESOURCE (MGR) ANNOTATION PROJECT
- MAIZE NUTRIGENOMICS
- MEDICINAL PLANT GENOMICS
- MEDICINAL PLANT TRANSCRIPTOMICS
- MINT GENOMICS
- ORYZA SNP PROJECT
- PACK-MULES AND GENOME EVOLUTION IN RICE
- PLANT REPEAT DATABASE
- PLANT RESILIENCE INSTITUTE
- POTATO 2.0
- POTATO GENOME VARIATION
- PYTHIUM GENOMICS
- RICE GENOME ANNOTATION PROJECT
- SINGLE CELL RNA SEQUENCING IN PLANTS
- SOLANACEAE COORDINATED AGRICULTURAL PROJECT
- SWEETGAINS
- SWITCHGRASS GENETIC DIVERSITY